Abstract
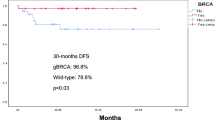
We describe the status and frequency of germline DNA genetic findings in an unselected prospective cohort of triple negative breast cancer patients participating in a platinum-based neoadjuvant chemotherapy trial. Study population includes 124 consecutive patients with stage II-III TNBC from a trial exploring the antitumor activity of neoadjuvant carboplatin/docetaxel chemotherapy enrolled between 2012 and March 2015, to determine the frequency of germline DNA genetic mutations. 17.1 % of the patients with germline DNA tested had deleterious mutations in any of the analyzed genes (12.38 % in BRCA1, 1.9 % in BRCA2 and BARD1 and 0.95 % in RAD51D). Attending the intrinsic subtype, all the BRCA1/2 carriers tested had basal-like subtype. Among wild-type (WT) patients, 70.11 % had basal subtype, 16.09 % HER2 enriched, 1.15 % Luminal B, and 4.60 % Normal-like. Mean age at diagnosis was significantly lower in mutation-carriers compared with no carriers (43.72 vs 53.10, p = 0.004). 3 BRCA1/2 carriers were detected between 51 and 60 years, and only one deleterious mutation (BARD1) over 60 years. A positive familiar history of breast and ovarian cancer was more frequent in patients with deleterious mutations (39.39 vs 17.94 %, p = 0.043). Our study confirms the prevalence of BRCA1/2 mutations in TNBC patients. TNBC should therefore be considered by itself as a criterion for BRCA1/2 genetic testing. Determination of other breast cancer predisposition genes implicated in homologous recombination should also be discussed in this population. However, no definitive conclusions can be reached due to the low prevalence and the uncertain clinical impact of most of the genes included.
Similar content being viewed by others
Change history
18 July 2017
An erratum to this article has been published.
Abbreviations
- BC:
-
Breast cancer
- ER:
-
Estrogen receptor
- FFPE:
-
Formalin-fixed, paraffin-embedded
- gDNA:
-
Genomic DNA
- HER2:
-
Human epidermal growth factor receptor 2
- HR:
-
Homologous recombination
- PR:
-
Progesterone receptor
- TNBC:
-
Triple-negative breast cancer
- VUS:
-
Variants of unknown significance
- WT:
-
Wild-type
References
Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA et al (2007) Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 13(15):4429–4434
Foulkes WD, Smith IE, Reis-Filho JS (2010) Triple-negative breast cancer. N Engl J Med 363(20):1938–1948
Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K et al (2006) Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA Am Med Assoc 295(21):2492–2502
Foulkes WD, Stefansson IM, Chappuis PO, Bégin LR, Goffin JR, Wong N et al (2003) Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 95(19):1482–1485
Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A et al (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100(14):8418–8423
Foulkes WD, Shuen AY (2013) In Brief: BRCA1 and BRCA2. J Pathol 230:347–349
Antoniou A, Pharoah PDP, Narod S, Risch HA, Eyfjord JE, Hopper JL et al (2003) Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72(5):1117–1130
Chen S, Parmigiani G (2007) Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol 25(11):1329–1333
Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkäs K, Roberts J et al (2014) Breast-cancer risk in families with mutations in PALB2. N Engl J Med 371:497–506
Laduca H, Stuenkel AJ, Dolinsky JS, Keiles S, Tandy S, Pesaran T et al (2014) Utilization of multigene panels in hereditary cancer predisposition testing: analysis of more than 2,000 patients. Genet Med 16(11):830–837
National Comprehensive Cancer Network (NCCN) 2015 The NCCN clinical practice guidelines in oncology TM 2010. Genetic/familial high risk assessment: breast and ovarian v2.2015. www.nccn.com
Sharma P, Klemp JR, Kimler BF, Mahnken JD, Geier LJ, Khan QJ et al (2014) Germline BRCA mutation evaluation in a prospective triple-negative breast cancer registry: implications for hereditary breast and/or ovarian cancer syndrome testing. Breast Cancer Res Treat 145(3):707–714
Daly MB, Pilarski R, Axilbund JE, Buys SS, Crawford B, Friedman S et al (2014) Genetic/familial high-risk assessment: breast and ovarian, version 1.2014. J Natl Compr Cancer Netw 12(9):1326–1338
Llort G, Chirivella I, Morales R, Serrano R, Sanchez AB, Teulé A et al (2015) SEOM clinical guidelines in hereditary breast and ovarian cancer. Clin Transl Oncol 17:956–961
Domagala P, Jakubowska A, Jaworska-bieniek K, Kaczmarek K, Durda K, Kurlapska A et al (2015) Prevalence of germline mutations in genes engaged in DNA damage repair by homologous recombination in patients with triple-negative and hereditary non- triple-negative breast cancers. PLoS One 2(6):1–14
Senkus E, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rutgers E et al (2015) Primary breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 26(Supplement 5):v8–v30
NCT 01560663. Predictors of response to neoadjuvant docetaxel-carboplatin chemotherapy for patients with stage II and III triple negative breast cancer. https://clinicaltrials.gov/ct2/results?term=NCT+01560663&Search=Search
Wolff AC, Hammond MEH, Hicks DG, Dowsett M, McShane LM, Allison KH et al (2013) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/College of American pathologists clinical practice guideline update. J Clin Oncol 31(31):3997–4013
Meindl A, Ditsch N, Kast K, Rhiem K, Schmutzler RK (2011) Hereditary breast and ovarian cancer: new genes, new treatments, new concepts. Dtsch Ärzteblatt Int. Deutscher Arzte-Verlag GmbH 108(19):323–30
Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER et al (2009) VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25(17):2283–2285
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9):1297–1303
Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D et al (2012) Ensembl 2012. Nucleic Acids Res 40:D84–D90
Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T et al (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol Off J Am Soc Clin Oncol 27(8):1160–1167
R Core Team (2015). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA et al (2000) Molecular portraits of human breast tumours. Nature 406(6797):747–752
Fostira F, Tsitlaidou M, Papadimitriou C, Pertesi M, Timotheadou E, Stavropoulou AV et al (2012) Prevalence of BRCA1 mutations among 403 women with triple-negative breast cancer: implications for genetic screening selection criteria: a Hellenic Cooperative Oncology Group Study. Breast Cancer Res Treat 134(1):353–362
Robertson L, Hanson H, Seal S, Warren-Perry M, Hughes D, Howell I et al (2012) BRCA1 testing should be offered to individuals with triple-negative breast cancer diagnosed below 50 years. Br J Cancer Nat Publ Group 106(6):1234–1238
Couch FJ, Hart SN, Sharma P, Toland AE, Wang X, Miron P et al (2015) Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol 33(4):304–311
Hartman AR, Kaldate RR, Sailer LM, Painter L, Grier CE, Endsley RR et al (2012) Prevalence of BRCA mutations in an unselected population of triple-negative breast cancer. Cancer 118:2787–2795
Wong-Brown MW, Meldrum CJ, Carpenter JE, Clarke CL, Narod SA, Jakubowska A et al (2015) Prevalence of BRCA1 and BRCA2 germline mutations in patients with triple-negative breast cancer. Breast Cancer Res Treat 150(1):71–80
Rummel S, Varner E, Shriver CD, Ellsworth RE (2013) Evaluation of BRCA1 mutations in an unselected patient population with triple-negative breast cancer. Breast Cancer Res Treat 137(1):119–125
Gonzalez-angulo AM, Timms KM, Liu S, Chen H, Jennifer K, Potter J et al (2012) NIH Public Access 17(5):1082–1089
Meyer P, Landgraf K, Högel B, Eiermann W, Ataseven B (2012) BRCA2 mutations and triple-negative breast cancer. PLoS One Public Library of Science 7(5):e38361
Muendlein A, Rohde BH, Gasser K, Haid A, Rauch S, Kinz E et al (2015) Evaluation of BRCA1/2 mutational status among German and Austrian women with triple-negative breast cancer. J Cancer Res Clin Oncol 141(11):2005–2012
Andrés R, Pajares I, Balmaña J, Llort G, Cajal TR, Chirivella I et al (2014) Association of BRCA1 germline mutations in young onset triple-negative breast cancer (TNBC). Clin Transl Oncol 16:280–284
Young SR, Pilarski RT, Donenberg T, Shapiro C, Hammond LS, Miller J et al (2009) The prevalence of BRCA1 mutations among young women with triple-negative breast cancer. BMC Cancer 9:86
Balmaña J, Díez O, Rubio IT, Cardoso F (2011) BRCA in breast cancer: ESMO Clinical Practice Guidelines. Ann Oncol Off J Eur Soc Med Oncol/ESMO 22(Suppl 6):vi31–vi34
De Brakeleer S, De Grève J, Desmedt C, Joris S, Sotiriou C, Piccart M et al (2016) Frequent incidence of BARD1-truncating mutations in germline DNA from triple-negative breast cancer patients. Clin Genet 89(3):336–340
Ollier M, Radosevic-Robin N, Kwiatkowski F, Ponelle F, Viala S, Privat M et al (2015) DNA repair genes implicated in triple negative familial non-BRCA1/2 breast cancer predisposition. Am J Cancer Res. 5(7):2113–2126
Eggington JM, Bowles KR, Moyes K, Manley S, Esterling L, Sizemore S et al (2014) A comprehensive laboratory-based program for classification of variants of uncertain significance in hereditary cancer genes. Clin Genet 86(3):229–237
Selkirk CG, Vogel KJ, Newlin AC, Weissman SM, Weiss SM, Wang C-H et al (2014) Cancer genetic testing panels for inherited cancer susceptibility: the clinical experience of a large adult genetics practice. Fam Cancer 13(4):527–536
Murray ML, Cerrato F, Bennett RL, Jarvik GP (2011) Follow-up of carriers of BRCA1 and BRCA2 variants of unknown significance: variant reclassification and surgical decisions. Genet Med 13(12):998–1005
Cheon JY, Mozersky J, Cook-Deegan R (2014) Variants of uncertain significance in BRCA: a harbinger of ethical and policy issues to come? Genome Med. 6(12):121
Mahon SM (2015) Management of patients with a genetic variant of unknown significance. Oncol Nurs Forum 42(3):316–318
Turner NC, Reis-Filho JS (2006) Basal-like breast cancer and the BRCA1 phenotype. Oncogene 25:5846–5853
Prat A, Adamo B, Cheang MCU, Anders CK, Carey LA, Perou CM (2013) Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 18(2):123–133
Zugazagoitia J, Pérez-Segura P, Manzano A, Blanco I, Vega A, Custodio A et al (2014) Limited family structure and triple-negative breast cancer (TNBC) subtype as predictors of BRCA mutations in a genetic counseling cohort of early-onset sporadic breast cancers. Breast Cancer Res Treat 148(2):415–421
Weitzel JN, Lagos VI, Cullinane CA, Gambol PJ, Culver JO, Blazer KR et al (2007) Limited family structure and BRCA gene mutation status in single cases of breast cancer. JAMA 297(23):2587–2595
Acknowledgments
This work was partially supported by the Ministry of Economy and Competitiveness ISCIII-FIS grants PI12/02684, and RD12/0036/0076, co-financed by ERDF (FEDER) Funds from the European Commission, “A way of making Europe.”
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The study was performed in accordance with the Declaration of Helsinki, approved by the ethics committees at all participating institutions and the Spanish Health Authority (Ethical Committee reference Area 1-Hospital General Universitario Gregorio Marañón).
Conflict of interests
The authors declare that they have no conflict of interest.
Informed consent
It was registered at www.clinicaltrials.gov (identifier code: NCT 01560663). The patients were diagnosed at any of the participant academic institutions including several hospitals from Spain and Peru. Informed consent was obtained from all individual participants included in this study.
Additional information
Milagros González-Rivera and Miriam Lobo have contributed equally to this work and should be considered as first authors as equal.
An erratum to this article is available at https://doi.org/10.1007/s10549-017-4396-0.
Rights and permissions
About this article
Cite this article
González-Rivera, M., Lobo, M., López-Tarruella, S. et al. Frequency of germline DNA genetic findings in an unselected prospective cohort of triple-negative breast cancer patients participating in a platinum-based neoadjuvant chemotherapy trial. Breast Cancer Res Treat 156, 507–515 (2016). https://doi.org/10.1007/s10549-016-3792-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-016-3792-1