Accelerate Your Oncology Research
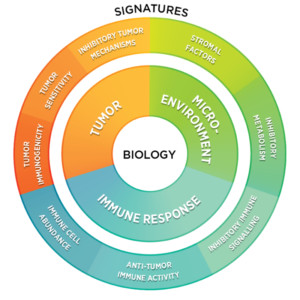
Cancer is a complex disease characterized by interactions between the tumor, microenvironment, and immune system. A broader and deeper understanding of the biology and signaling pathways that lead to cancer enables scientists to better identify, characterize, and target biomarkers that can ultimately be translated into clinical applications. Advances in molecular tools now enable a multiomic, holistic view of cancer that rapidly advances the understanding of how tumors develop, evade the immune system, and progress.
Challenges
Cancer is a constellation of highly heterogenous diseases that share a common origin in mutations that drive key cellular functions like proliferation and cell death. Each cell in a tumor can bear its own genomic alterations and expression programs. Tissue-based and microenvironmental pressures add their own unique features to tumor pathology. Patients may also respond differently to treatment due to their own genetic makeup and microbiome, creating challenges for translating insights from model organisms to humans. Finally, the immune response adds an additional axis of complexity that adds to the challenge of cancer research and treatment.
Cancer is not just one disease
Tumor microenvironment impacts immune response

Not all patients respond to treatments
Your Trusted Partner in Cancer Research
NanoString has been developing cancer research tools for over 20 years and believes in partnering with scientists to advance their research. We offer integrated, multiomic research tools for every stage of the translational research continuum. Whether you’re profiling the expression of 800+ genes with the nCounter® Analysis System, performing spatial profiling with the GeoMx® Digital Spatial Profiler (DSP), or getting single cell and subcellular spatial multiomics data with the CosMx® Spatial Molecular Imager (SMI), we are here to partner with you every step of the way.
The CosMx® SMI and decoder probes are not offered and/or delivered to the Federal Republic of Germany for use in the Federal Republic of Germany for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof in a method used in fluorescence in situ hybridization for detecting a plurality of analytes in a sample without the consent of the President and Fellows of Harvard College (Harvard Corporation) as owner of the German part of EP 2 794 928 B1. The use for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof is prohibited without the consent of the President and Fellows of Harvard College (Harvard Corporation).
Three Platforms. Unlimited Potential.
Overcome cancer heterogeneity with a suite of translational tools and solutions that empower you to get actionable insights faster at any scale, from single cells to multi-cellular molecular compartments in the tumor microenvironment.

nCounter®
Analysis System
- Discover predictive and prognostic biomarkers
- Evaluate mechanisms of treatment response
- Monitor disease biomarkers in clinical trials
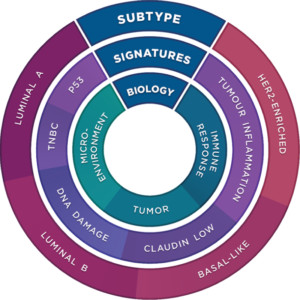
- Stratify patients using validated TIS, PAM50 and LST gene signatures
Select nCounter Oncology Panels
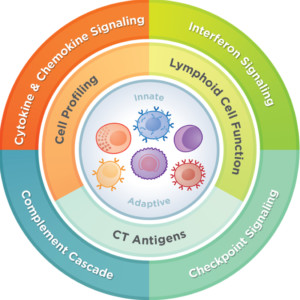
The nCounter Oncology portfolio offers a multitude of curated panels covering application areas in immuno-oncology, breast cancer research, cell and gene therapy, drug development and biomarker discovery, and so much more.
GeoMx DSP Assays for Oncology
CosMx SMI Assays for Oncology Research
Spotlight on Success
“The nCounter Analysis System, GeoMx DSP and CosMx SMI are highly complementary for oncology research, especially for biomarker discovery. nCounter, with its smartly curated panels, allows for rapid screening of alterations in gene expression as well as insight into pathway activity. GeoMx reveals the regions of tissue where these alterations occur, and CosMx deepens this insight down to the single cell and subcellular level. This powerful instrument combo provides an unprecedented view of molecular tissue architecture that can be applied to basic and translational research, especially in cases where tissues samples are scarce and precious.”

Nina Radosevic
Investigative Pathologist, Centre Jean Perrin, INSERM U 1240, Clermont-Ferrand, France
Related Resources



Case Studies and Testimonials
Oncology Customer Testimonials
Oncology Case Studies
Publications
SITC 2023: Gene signature predicts autoimmune toxicity in patients with metastatic melanoma
Expression patterns of microRNAs and associated target genes in ulcerated primary cutaneous melanoma
Neoadjuvant enoblituzumab in localized prostate cancer: a single-arm, phase 2 trial
B7 homolog 3 (B7-H3; CD276), a tumor-associated antigen and possible immune checkpoint, is highly expressed in prostate cancer (PCa) and is associated with early recurrence and metastasis. Enoblituzumab is a humanized, Fc-engineered, B7-H3-targeting antibody that mediates antibody-dependent cellular cytotoxicity.
Digital spatial profiling of intraductal papillary mucinous neoplasms: Toward a molecular framework for risk stratification
The histopathologic heterogeneity of intraductal papillary mucinous neoplasms (IPMN) complicates the prediction of pancreatic ductal adenocarcinoma (PDAC) risk. Intratumoral regions of pancreaticobiliary (PB), intestinal (INT), and gastric foveolar (GF) epithelium may occur with either low-grade dysplasia (LGD) or high-grade dysplasia (HGD).
Single-cell heterogeneity of EGFR and CDK4 co-amplification is linked to immune infiltration in glioblastoma
Glioblastoma (GBM) is the most aggressive brain tumor, with a median survival of ∼15 months. Targeted approaches have not been successful in this tumor type due to the large extent of intratumor heterogeneity.

Contact Us
Have questions or simply want to learn more?
Contact our helpful experts and we’ll be in touch soon.