nCounter® Breast Cancer 360™ Panel
Helping Your Research
Want a system that provides a unique 360° view of gene expression for the breast tumor, microenvironment, and immune response? If yes, then you have found the right product. The Human nCounter Breast Cancer 360 panel and data analysis report helps researchers quickly decode the complexities of breast cancer biology, develop novel breast cancer gene signatures, and categorize disease heterogeneity using 48 biological signatures including signatures based upon the validated PAM50 and Tumor Inflammation Signature (TIS) assays. Get the most out of your panel by quickly distilling a large amount of data into actionable signatures measuring variables crucial to breast tumor-immune interaction.
Inspired by systems biology approaches to cancer research, NanoString’s 360 Series Panel Collection gives you a 360° view of gene expression by combining carefully-curated content involved in the biology of the tumor, microenvironment, and the immune response into a single holistic assay. Each panel contains the 18-gene Tumor Inflammation Signature (TIS) that measures a peripherally-suppressed, adaptive immune response and has been shown to correlate with response to checkpoint inhibitors.

Panel Selection Tool
Find the gene expression panel for your research with Panel Pro
Find Your Panel
Product Information
Includes expertly curated 776 genes across 23 categories of breast cancer tumor biology to support the evaluation of pathways and process, as well as novel signature development.
Content included in the Breast Cancer 360 panel allows for a more comprehensive measurement of biological variables crucial to tumor progression and response to a wide range of treatments. Research signatures are enriched with potentially predictive genes involved in proliferation, endothelium, angiogenesis, cytotoxicity, stroma, inflammatory chemokines, and apoptosis.
- 48 signatures including two analytically validated signatures- PAM50 and Tumor Inflammation Signature
- 10 research-focused signatures and 30 novel signatures measuring important tumor and immune activities
- adapted to decode breast cancer biology in concert
Analytically Validated Signatures
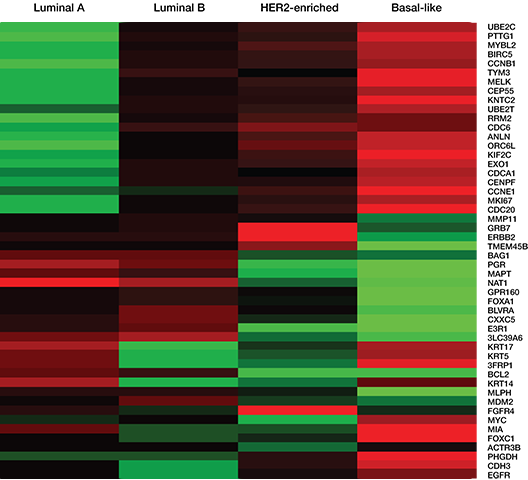
PAM50 Signature1,2
Included with the Breast Cancer 360 panel is the PAM50 Signature. This 50-gene signature measures a gene expression profile that allows for the classification of breast cancer into four biologically distinct subtypes and a prognostic score.
- PAM50 Subtype
- Luminal A
- Luminal B
- HER2-Enriched
- Basal-like
- Prosigna Score / Risk of Recurrence
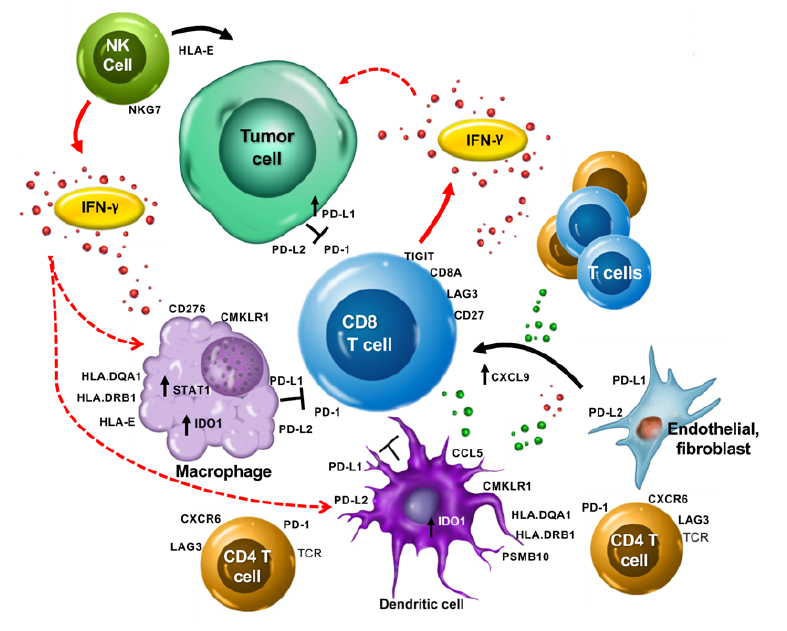
Tumor Inflammation Signature3
Included with the Breast Cancer 360 panel is the Tumor Inflammation Signature. This 18-gene signature measures activity known to be associated with response to PD-1/PD-L1 inhibitors pathway blockade3.
- Includes 4 areas of immune biology used to determine peripherally suppressed immune response and the identification of “hot” or “cold” tumors
- Antigen Presenting Cells
- T Cell/NK presence
- IFNγ Biology
- T Cell Exhaustion
- Tissue-of-origin agnostic (Pan-cancer)
- Potential surrogate for PD-L1 and mutational load in research setting4
View publication and video.
360 Series Product Comparison
Fully-annotated gene lists in Excel format are available for each of the 360 Panels. The table below compares the biology coverage of the 360 Panels across the tumor, microenvironment, and the immune response to that of the PanCancer Panels Collection.
Related Resources
Publications
Assessing Longitudinal Treatment Efficacies and Alterations in Molecular Markers Associated with Glutamatergic Signaling and Immune Checkpoint Inhibitors in a Spontaneous Melanoma Mouse Model
Previous work done by our laboratory described the use of an immunocompetent spontaneous melanoma-prone mouse model, TGS (TG-3/SKH-1), to evaluate treatment outcomes using inhibitors of glutamatergic signaling and immune checkpoint for 18 weeks. We showed a significant therapeutic efficacy with a notable sex-biased response in male mice.
Spatial transcriptomics reveals discrete tumour microenvironments and autocrine loops within ovarian cancer subclones
High-grade serous ovarian carcinoma (HGSOC) is genetically unstable and characterised by the presence of subclones with distinct genotypes. Intratumoural heterogeneity is linked to recurrence, chemotherapy resistance, and poor prognosis.
Spatially Segregated Macrophage Populations Predict Distinct Outcomes In Colon Cancer
Tumor-associated macrophages are transcriptionally heterogeneous, but the spatial distribution and cell interactions that shape macrophage tissue roles remain poorly characterized. Here, we spatially resolve five distinct human macrophage populations in normal and malignant human breast and colon tissue and reveal their cellular associations.
Request a Quote
Contact our helpful experts and we’ll be in touch soon.