CosMx® Spatial Molecular Imager
Elevate your single-cell research with CosMx® SMI, the high fidelity Spatial Single-Cell Imaging Platform.
CosMx SMI is the only in situ imaging platform to offer whole transcriptome single-cell spatial biology with unmatched accuracy, sensitivity, and genomic breadth. It provides spatial multiomics with formalin-fixed paraffin-embedded (FFPE) and fresh frozen (FF) tissue samples at single cell and subcellular resolution.
CosMx SMI enables rapid quantification and visualization of the entire whole transcriptome and 64 validated proteins, delivering best-in-class performance for the discovery of unknown and unexpected biology. Capture the story of every cell-to-cell interaction from an entire tissue section with this industry leading, spatial single-cell imaging platform, and drive deeper insights for cell atlasing, tissue phenotyping, cell-cell interactions, cellular processes, and biomarker discovery.
Resolve millions of cells and billions of transcripts at single-cell and subcellular resolution
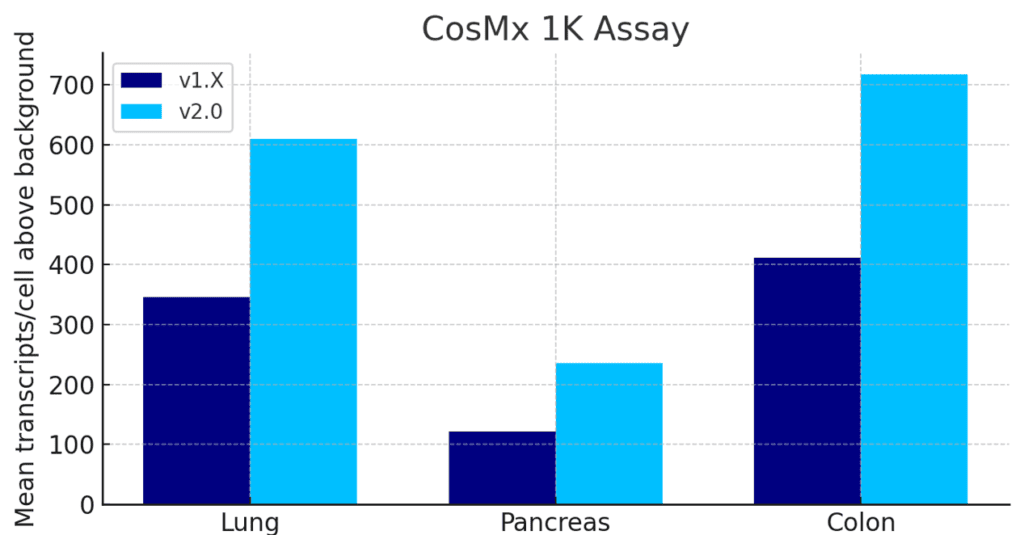
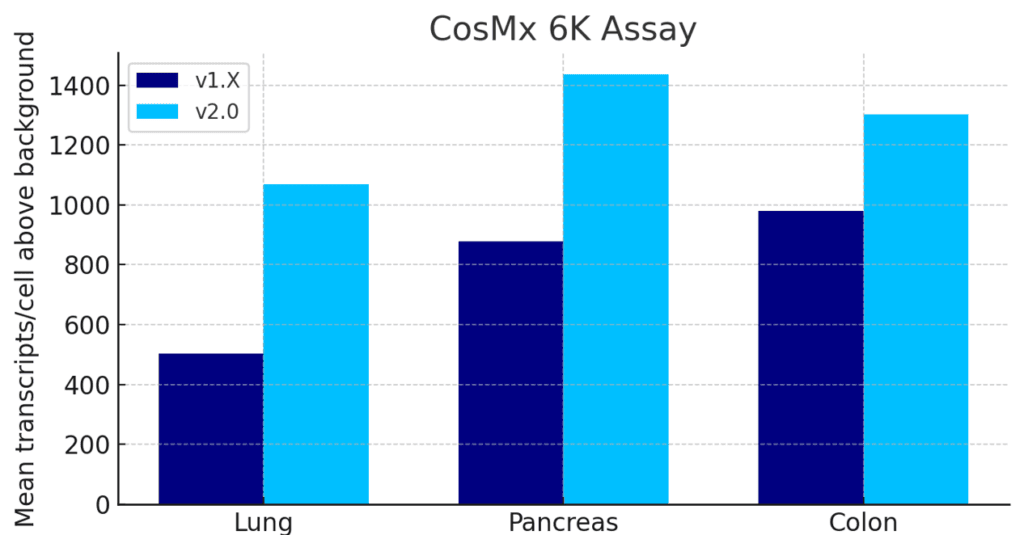
Unmatched Single Cell Spatial Sensitivity and Cell-Segmentation with CosMx 2.0
The latest software upgrade for CosMx (v2.0) delivers 1.5 to 2x sensitivity increases and best in class cell segmentation across all assays.
The CosMx® SMI and decoder probes are not offered and/or delivered to the Federal Republic of Germany for use in the Federal Republic of Germany for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof in a method used in fluorescence in situ hybridization for detecting a plurality of analytes in a sample without the consent of the President and Fellows of Harvard College (Harvard Corporation) as owner of the German part of EP 2 794 928 B1. The use for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof is prohibited without the consent of the President and Fellows of Harvard College (Harvard Corporation).



“We are impressed with ease of use with which the CosMx SMI enabled single cell spatial transcriptomics across various tissue types, even in archival FFPE tissue. The high-plex protein assay makes the CosMx SMI a true multiomic solution. The CosMx SMI’s large capture area allows analysis from valuable clinical trial samples using tissue microarrays, enabling characterization of 100s of samples weekly.”

Dr. Nigel B Jamieson
Professor of Surgery, University of Glasgow, UK
“At last count, we have processed 160 samples with the CosMx SMI, including tumor and mouse brain samples. This technology uncovers vital insights into cell biology, enhancing our understanding of both malignant and normal tissues. It is crucial for creating spatial biomarkers and improving patient outcomes. Plus, the equipment and protocols are reliable and user-friendly.”

Dr. Aubrey Thompson
Mayo Clinic Comprehensive Cancer Center, Florida
“The CosMx SMI complemented our efforts to generate organ atlases of the healthy human system together with our partners of the Human Cell Atlas project. It allowed us to visualize single-cell reference maps directly in organs. The scale of 1000-plex gene panel with image-guided cell segmentation makes the CosMx SMI, a unique platform for spatial genomics!”

Dr. Holger Heyn
Team Leader, Single Cell Genomics Group, CNAG, Spain
“The CosMx SMI instrument has become integral to our laboratory’s future direction, and the responsiveness and skill of the customer support for it and the AtoMx SIP has been absolutely fantastic.”

Dr. Grant Kolar
Director, Research Microscopy and Histology Core, St. Louis University School of Medicine
Be the first to get…
Comprehensive answers with over 18,000 plex RNA assays in a single experiment


6K Discovery 6175-plex, Human
Whole Transcriptome ~19,000-plex, Human
Get single-cell gene expression in challenging FFPE tissues with spatial context.
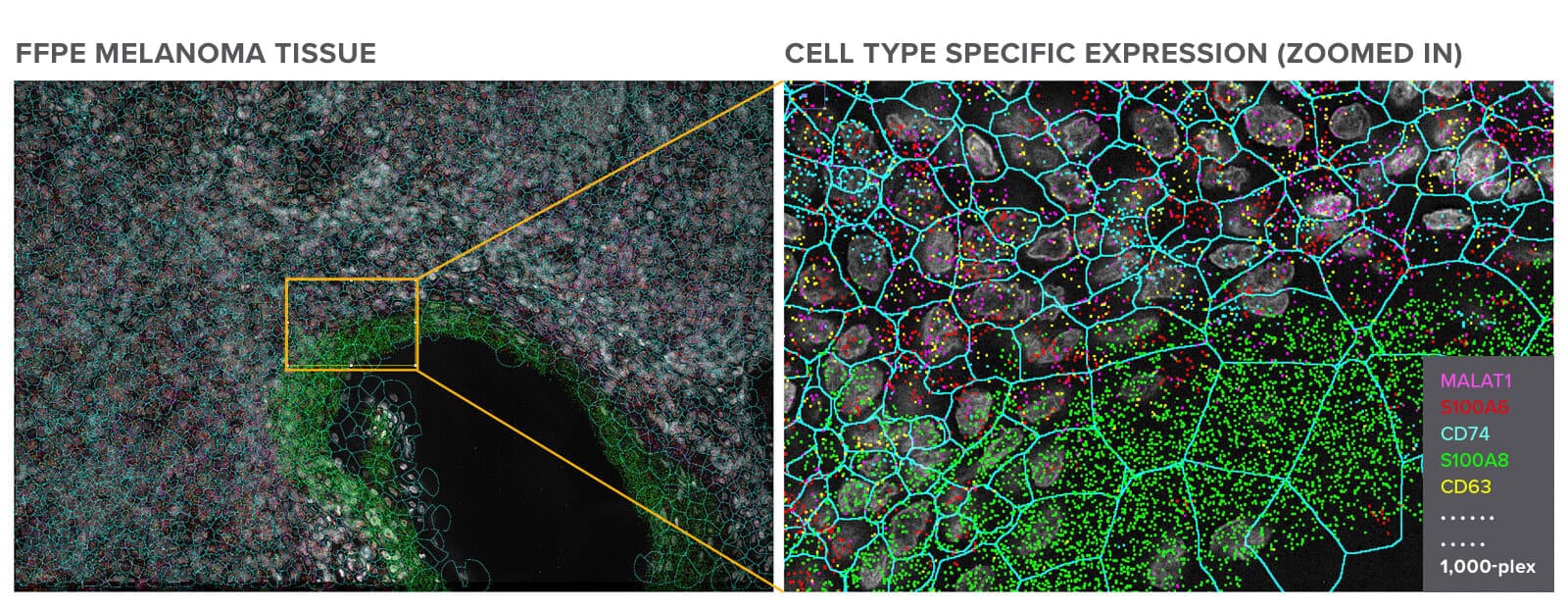
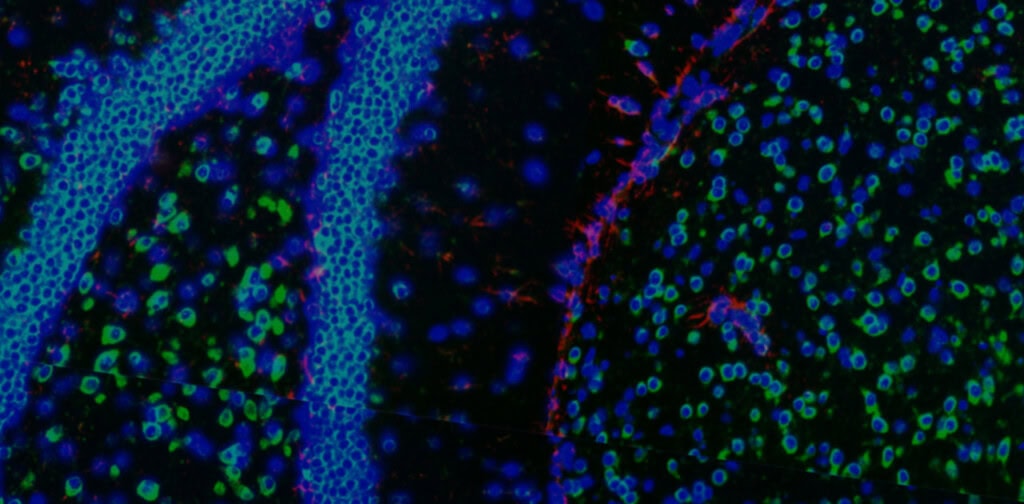
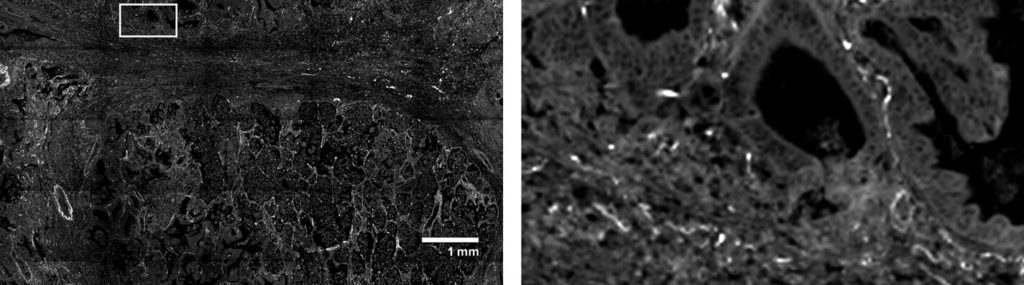
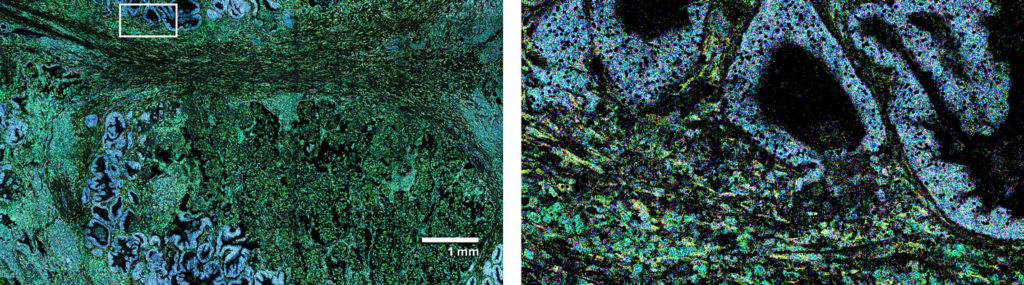
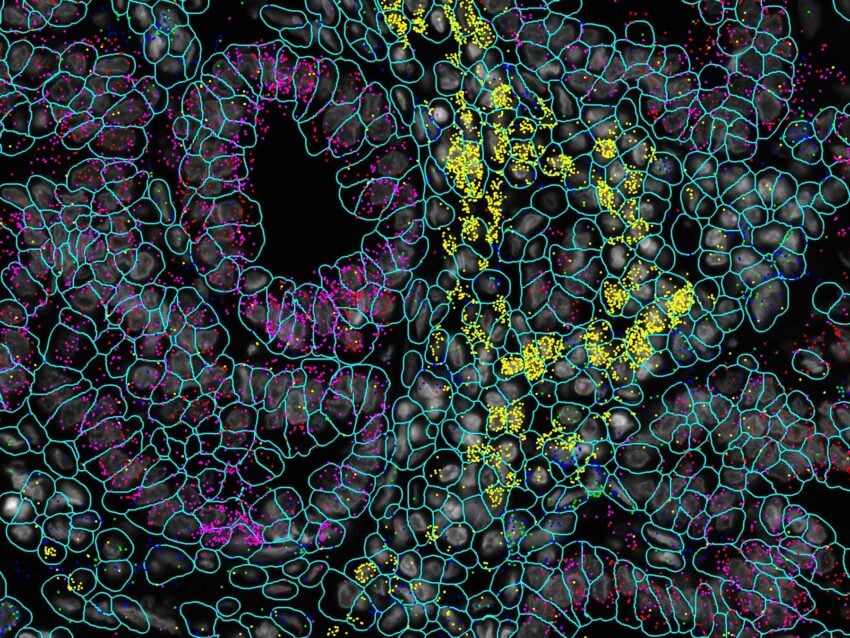
Melanoma FFPE tissue probed with 1000-plex RNA panel to detect spatial localization of transcripts in intact tissue.
CosMx SMI delivers the highest level of sensitivity, capturing the largest number of transcripts per cell and unique genes per cell versus all other spatial imaging platforms on FFPE samples.
Total transcripts per cell
Unique genes per cell
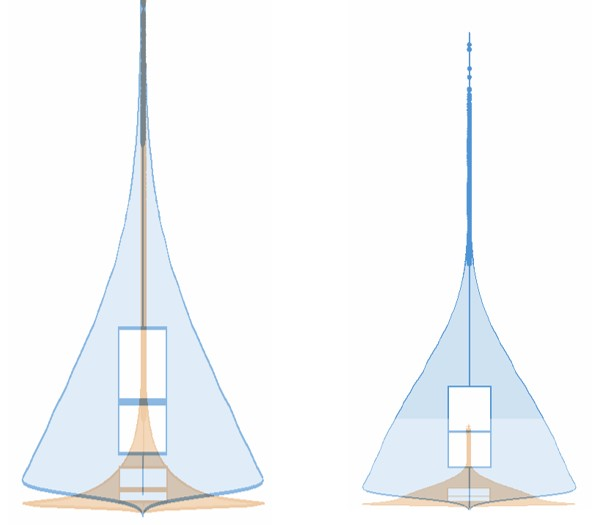
Example: multiple serial section comparative study
CosMx SMI
Other
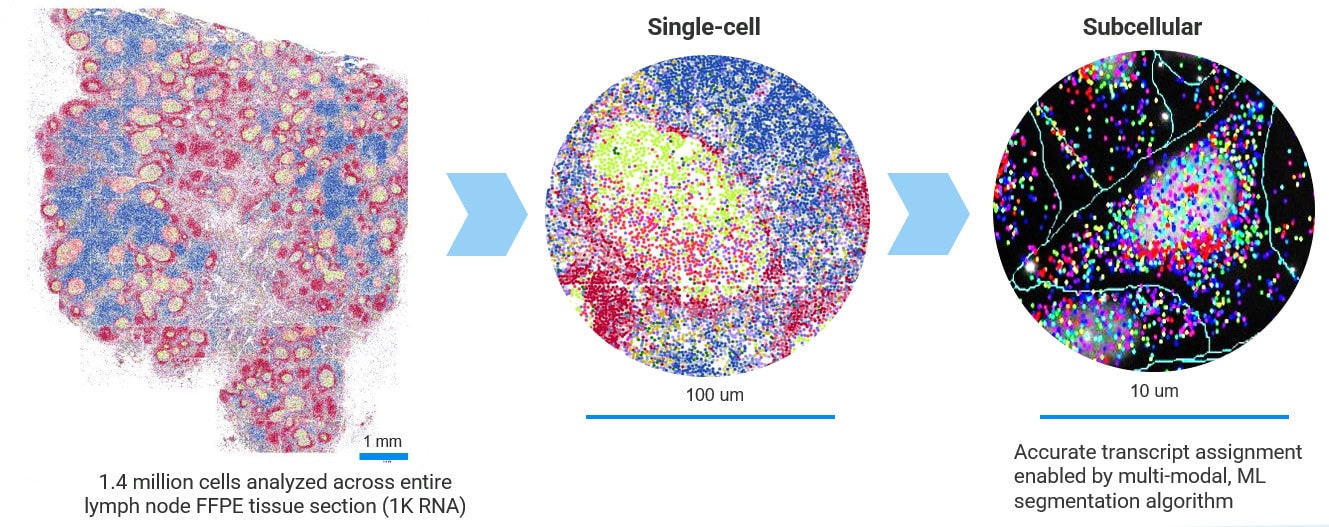
Multi-modal approach for true single-cell segmentation
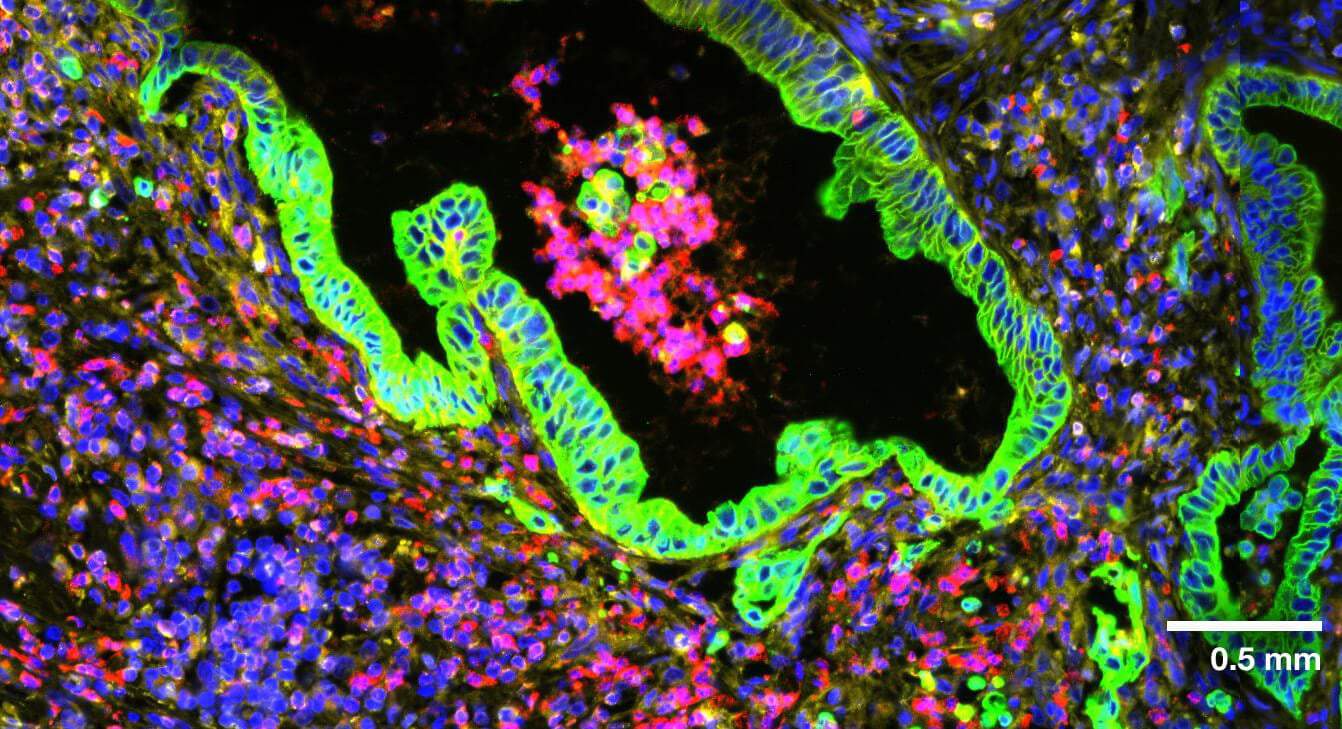

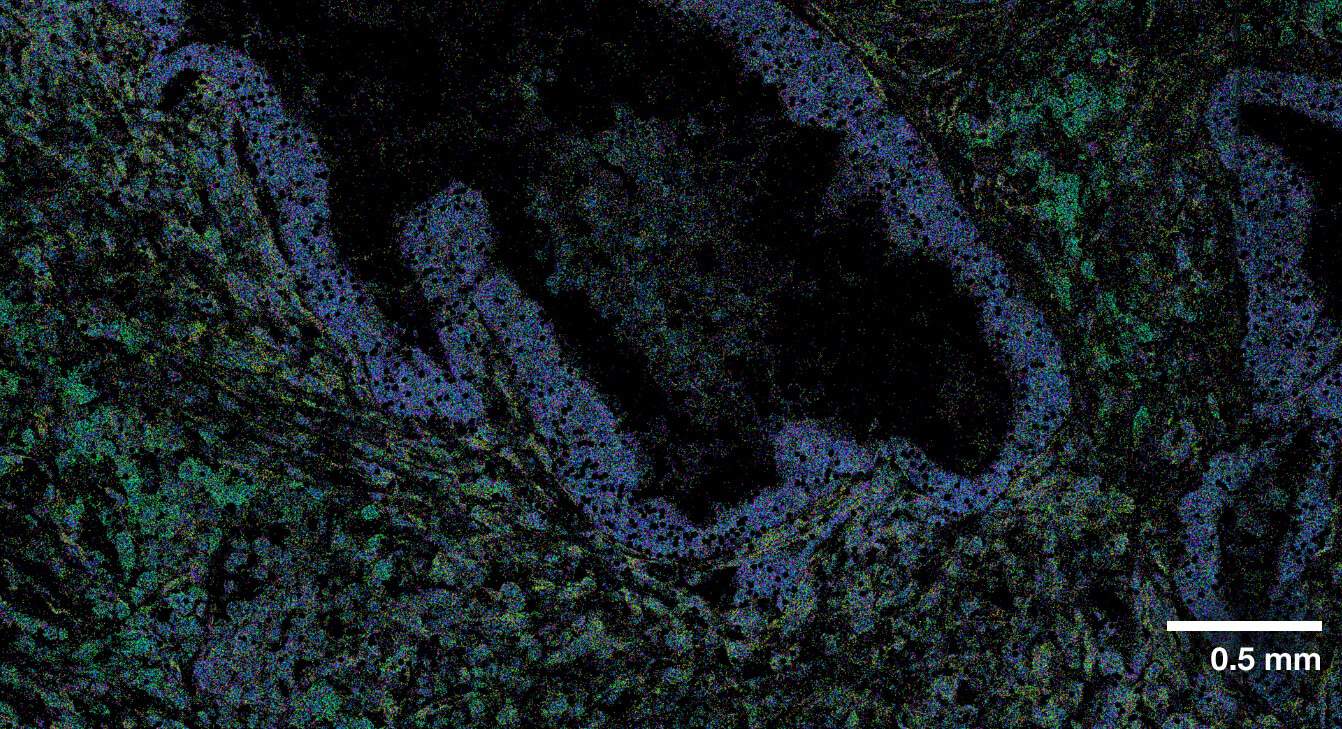
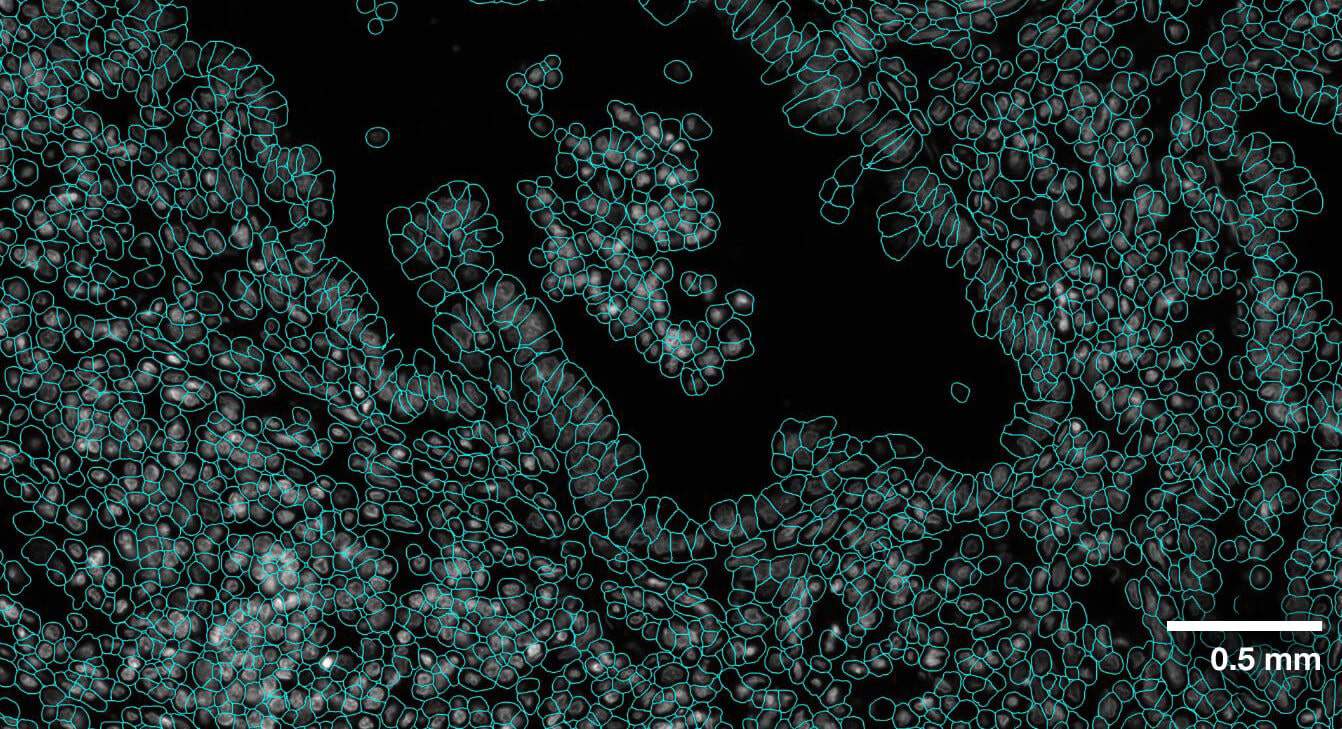
Multi-modal cell segmentation process provides accurate cell boundaries detection. CosMx cell segmentation uses cell membrane and morphology marker protein images, machine-learning augmented cell segmentation algorithm and transcript-based segmentation refinement to achieve precise single-cell segmentation in morphologically intact tissue.
CosMx Same-Cell Multiomics with RNA and Protein for Unlimited Spatial Exploration

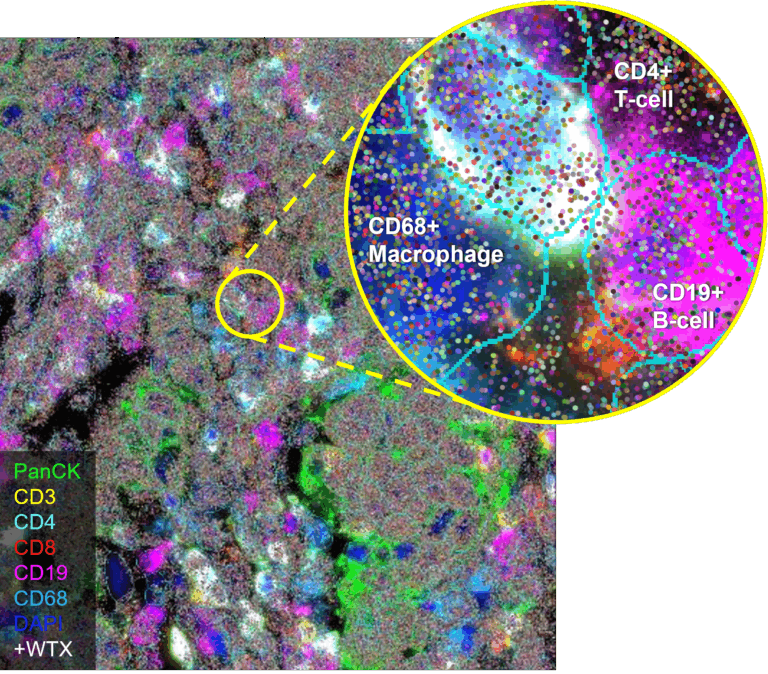
Identify cell types and states with greater accuracy by combining transcriptomic profiles with protein expression in the same cell.
More info
CosMx SMI vs. Xenium: Comparative Analysis of Breast Cancer Tissue Samples
How it works
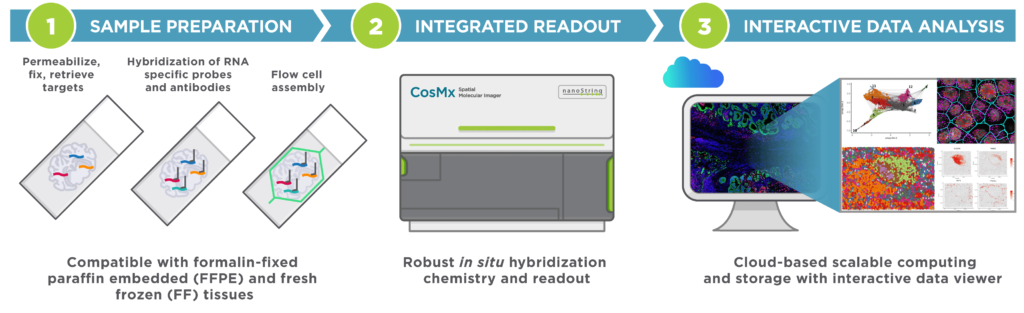
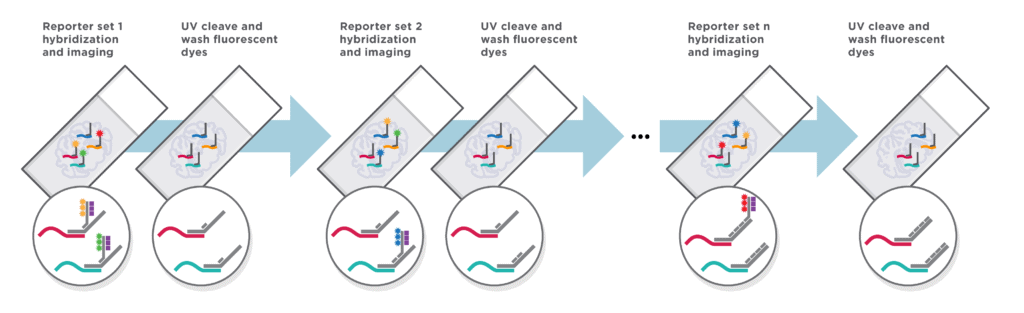
CosMx SMI is an integrated system with mature cyclic fluorescent in situ hybridization (FISH) chemistry, high-resolution imaging readout, interactive data analysis and visualization software.
Easy Sample Preparation, Compatible with Any Sample Type
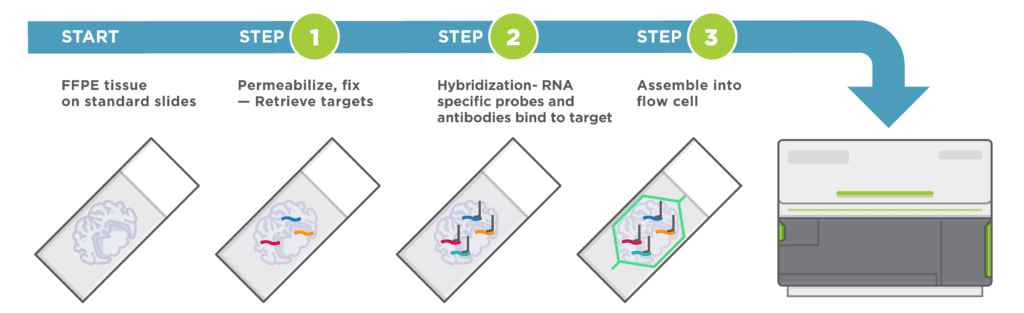
Automated Cyclic in situ Hybridization Chemistry
Applications
CosMx Spatial Molecular Imager is the most flexible and robust spatial single-cell imaging platform for:
- Cell Atlas and Characterization: Define cell types, cell states, tissue microenvironment phenotypes, and gene expression networks within spatial context.
- Cell-cell Interaction: Understand biological process controlled by ligand-receptor interactions.
- Spatial Biomarkers: Quantify change in gene expression based on treatment and identify single-cell subcellular biomarkers with spatial context.

Interested in learning more about CosMx SMI for Single-Cell Imaging?
Contact our helpful experts and we’ll be in touch soon.

Related Resources




Support Documents
Single-Cell Imaging FAQs
Why is a single cell important?
Cells are a fundamental unit of life. A comprehensive understanding of how cells organize themselves in different layers of information to form tissues is not yet fully achieved. Further, no matter how seemingly homogeneous a tissue might appear, it contains a diverse population of cells, all of which represent different manifestations of that tissue type. Learn more »
Why is single-cell analysis important?
Single-cell analysis encompasses the study of genomics, transcriptomics, proteomics, and metabolomics at single-cell resolution. As cells are the organism’s building blocks, they are organized in different layers of information to form tissues, and the position of each cell within a tissue has a physiological or morphological function. Learn more »
What is single-cell technique?
Single-cell techniques are advances in single-cell manipulation and amplification that have enabled the study of genomics, transcriptomics, and epigenomics at the level of a single cell. Learn more »
What is single-cell spatial transcriptomics?
Analysis of mRNA expression profile with spatial context at the level of a single cell is known as single-cell spatial transcriptomics. Each cell has a unique transcriptomic fingerprint as gene expression patterns can be heterogeneous even amongst similar cells in both standard and abnormal cell states. Learn more »
Is spatial transcriptomics single-cell resolution?
Yes. Recent advances in the field of spatial transcriptomics have made it possible to visualize RNA transcripts at the resolution of a single-cell and, in some cases, subcellular resolution. Learn more »
What is single-cell analysis used for?
Single-cell analysis can provide data on cellular phenotypes by studying the effects of genomic alterations, gene expression, and environmental influences at the level of a single cell. Learn more »