CosMx™ SMI Datasets
See More Spatial Biology from Precious Samples
CosMx™ Spatial Molecular Imager (shipping now) is the first fully-integrated multiomics single-cell spatial biology solution and enables researchers to investigate morphologically-intact tissues at unprecedented plex, resolution and cell numbers. CosMx SMI enables imaging of more than 1,000 RNA and over 64 protein analytes at subcellular resolutions across more than 1 million cells within both fresh frozen and formalin-fixed paraffin-embedded (FFPE) tissue samples.
The CosMx™ SMI and decoder probes are not offered and/or delivered to the Federal Republic of Germany for use in the Federal Republic of Germany for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof in a method used in fluorescence in situ hybridization for detecting a plurality of analytes in a sample without the consent of the President and Fellows of Harvard College (Harvard Corporation) as owner of the German part of EP 2 794 928 B1. The use for the detection of cellular RNA, messenger RNA, microRNA, ribosomal RNA and any combinations thereof is prohibited without the consent of the President and Fellows of Harvard College (Harvard Corporation).
Analyzing RNA and protein in FFPE samples is known to be challenging due to their limited quantities, sample degradation and workflows required to work with them. The CosMx SMI system leverages simple sample preparation workflows and robust in situ hybridization chemistry and works with any sample type.
We have generated open-source datasets for FFPE samples to highlight the power of CosMx SMI. CosMx SMI combines ultimate sensitivity, specificity, robust cell segmentation and broad, high plex panels to enable researchers to explore the cell atlas, cell-cell interactions and phenotypes of the tissue microenvironment.
Available Datasets

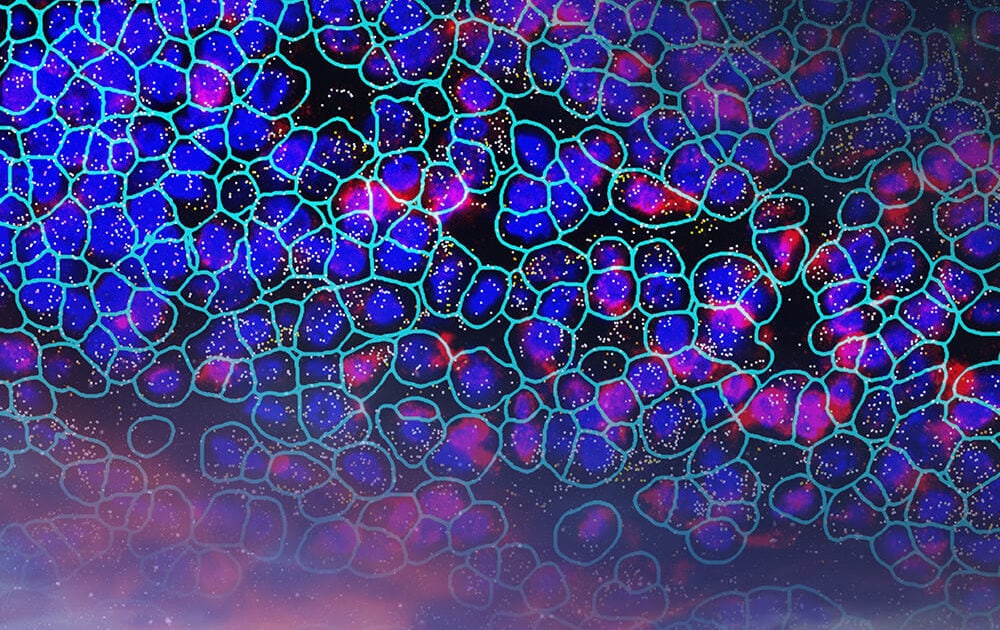
Human Pancreas (RNA)
Elevate to the ultimate Plex with CosMx Whole Transcriptome Panel. Explore the human pancreas dataset which can identify rare cell types, and understand cell-to-cell interactions in spatial context, providing a comprehensive understanding of cellular landscapes that was previously inaccessible.
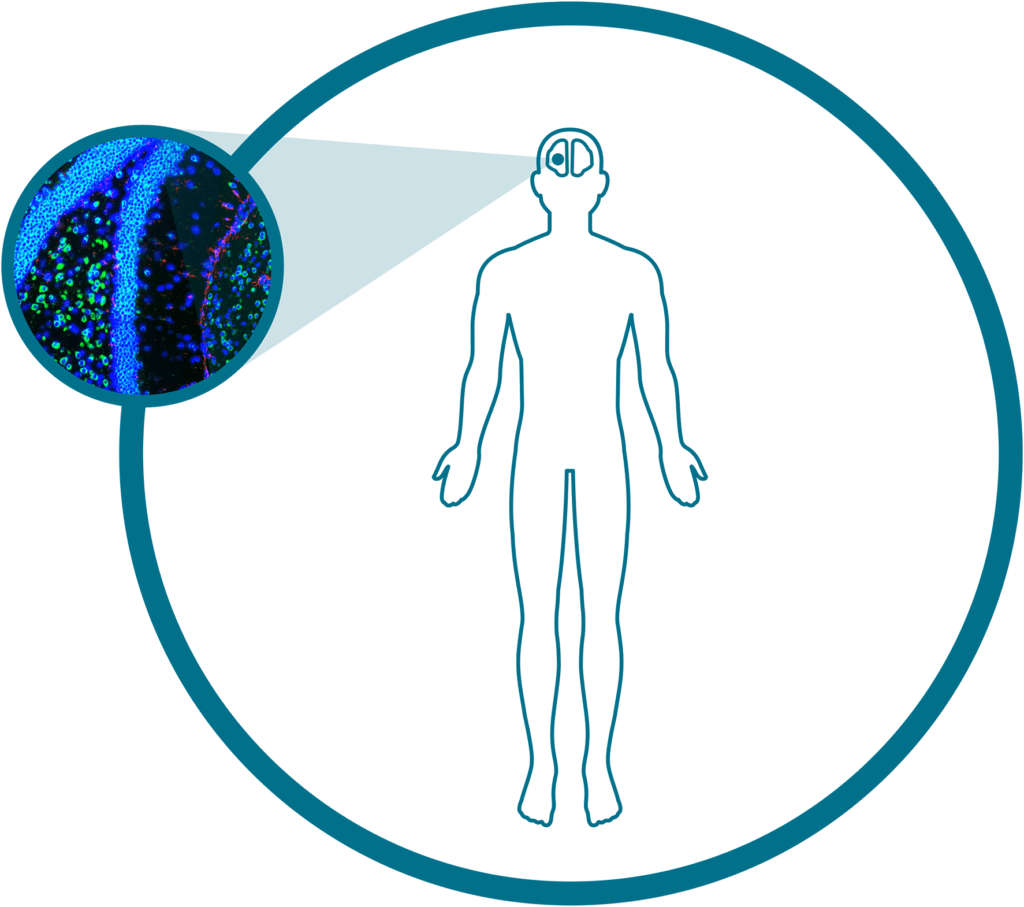
Human Frontal Cortex (RNA)
Make the leap to single-cell spatial with this open-source dataset on human brain tissue showcasing over 6000 genes. This dataset from a ~100mm2 region of human frontal cortex totals over 194,000 single cells and ~331 million transcripts.
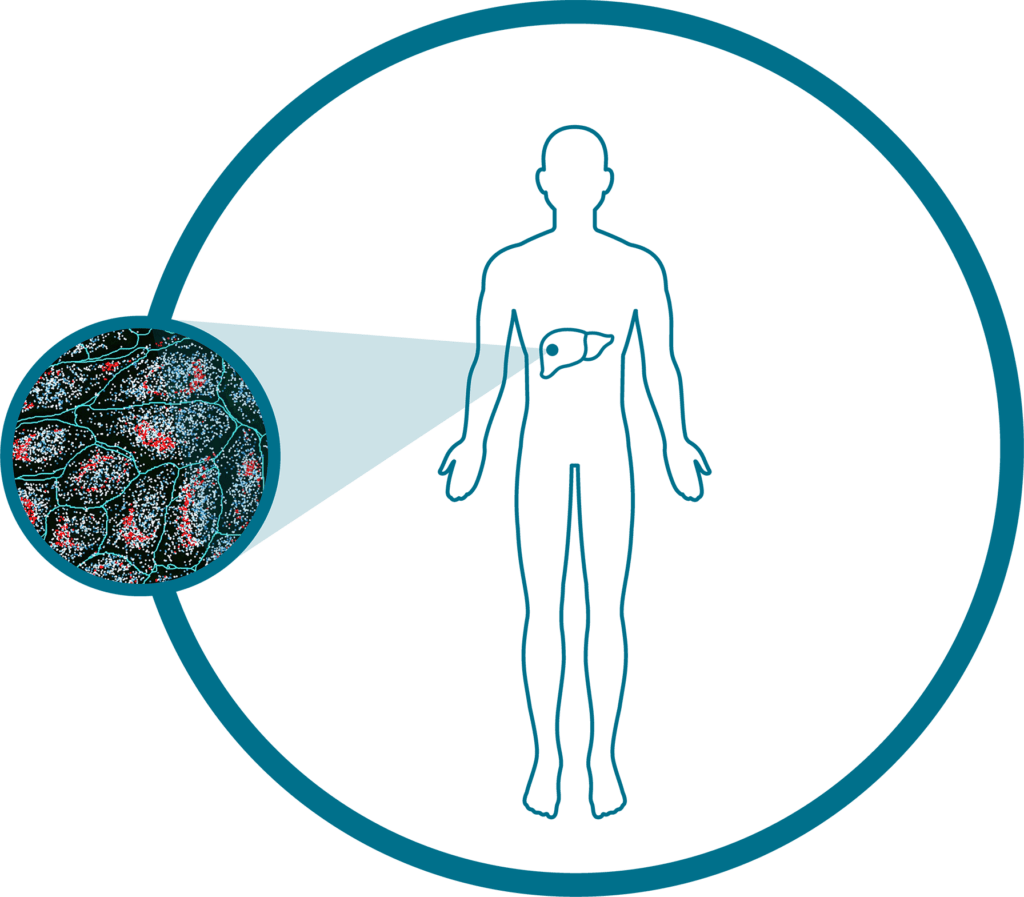
Human Liver (RNA)
Explore spatially resolved single-cell data from FFPE samples of normal and cancerous liver tissue to reveal spatial insights into the disease state, TME, protein imaging-based cell segmentation, a subcellular expression map of 1000+ genes, and a spatially-resolved cell type map.

Mouse Brain (RNA)
Explore mouse brain FFPE tissue across two coronal sections, one covering a full hemisphere and the other spanning the hippocampal formation and cortex bilaterally, totalling over 90,000 single cells and ~225 million transcripts.
We also have an archive of a Non-Small Cell Lung Cancer dataset — done on a CosMx™ SMI prototype — available for review.