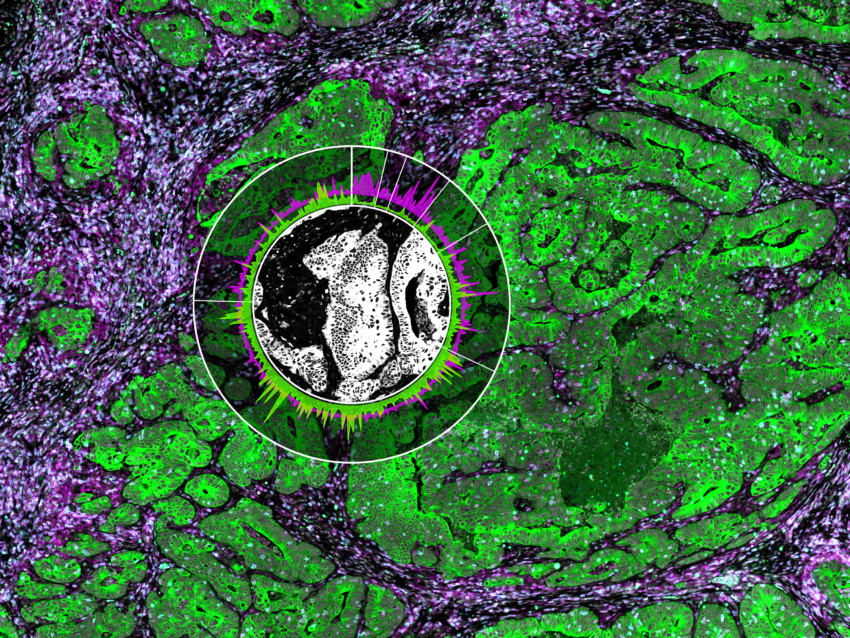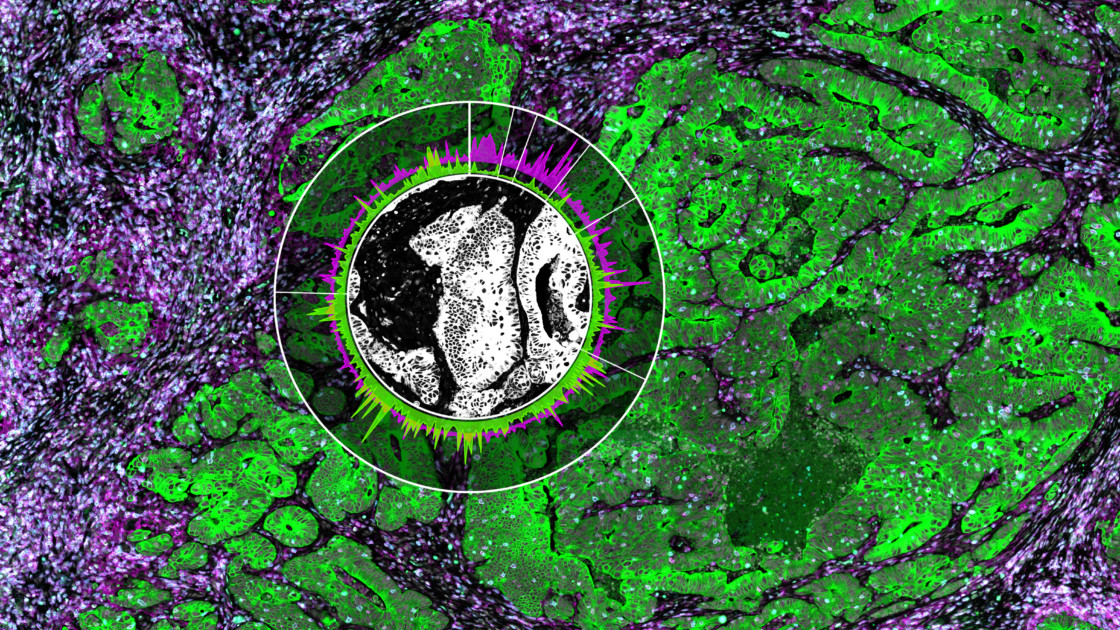
Why Spatial Biology Enhances Spatial Transcriptomics Data
Spatial Biology: this term carries an exciting space odyssey-like feel, although its processes take place on planet Earth. Even so, the field of spatial biology is by no means less thrilling and pioneering than the outer reaches of space.
Understanding Spatial Biology
Spatial Biology Definition
Spatial Biology is defined as the study of tissues within their own 2D or 3D context. It is the new frontier of molecular biology.
Consider how GPS captures location coordinates within an area to create a map and track you within it. The same principle can be translated to spatial data on a cellular and molecular level.
Through this technology, we can help map the spatial architecture of a cell and how it talks to and interacts with its surroundings. Spatial biology is like being inside a tissue sample at a molecular level. It enables single-cell analysis. Through spatial biology tools, you are empowered to see things that are not possible by sequencing or any other technologies out there.
The NanoString GeoMx® Digital Spatial Profiler (DSP) makes spatial genomics and spatial transcriptomics in the life sciences industry possible. It uses software to visualize the spatial architecture for image analysis and analyze multiple biomarkers of a tissue sample.
Spatial Biology In Practice
An increasing number of scientists are finding spatial biology extremely useful for studying oncology, immune-oncology, neurobiology, and even COVID research. Check out this link on how other scientists are using spatial biology for COVID research.
Single-cell genomics and microscopic analysis can provide information on genes and protein expression. However, even with spatial discrimination, these technologies have important pitfalls:
- Gene analysis on microdissected or dissociated tissues. Gene expression from laser capture microdissection or single cells can be informative. But, the context of interactions with surrounding cells is lost.
- In situ hybridization or standard immunofluorescence. These techniques enable the detection and localization of nucleic acid sequences or proteins in fixed tissue but have severely limited multiplexing capabilities. They also may suffer from quantitation challenges or extreme non-linearities depending on the method of visualization.
GeoMx DSP has solved both issues, with the added value of being enhanced for the hardest and yet most common sample type of all: formalin-fixed paraffin-embedded (FFPE).
Why do Spatial Omics Matter?
Nature Methods dubbed spatial transcriptomics “method of the year” for 2020. Its explosive popularity and potential for future insight made it a clear top choice.
In the words of Nature Methods staff: “spatially resolved transcriptomics highlights how these technologies have matured and expanded to give biologists exceptional views of the biology of single cells while retaining information on spatial context.”
Why is GeoMx DSP the Spatial Biology Technology of the Future?
The key breakthrough with the GeoMx DSP technology is a spatial approach. This results in the whole biology world of microscopy being turned upside down.
In typical experiments, a scientist adds detection reagents directly to the tissue and reads the results out with a microscope. Through NanoString’s innovative approach, detection is done in a much different manner.
First, each affinity reagent molecule binds to its intended target, which is linked to a unique DNA sequence with a UV light-cleavable linker.
Next, these are placed on the tissue section(s) of interest, effectively “staining” them invisibly.
Then, the focused UV light from the microscope liberates the indexing oligonucleotides from a region of interest (ROI). These ROIs are user-defined with the software and released into the liquid above the slide. They are collected via a thin tube and stored in a plate for subsequent quantitation.
This is repeated for each region of interest to map protein or transcriptional activity within the tissue architecture.
The spatially resolved pools of oligonucleotides are hybridized to fluorescent barcodes. As a result, this enables the digital counting of up to ~1 million individual targets per ROI.
This is done by using the standard NanoString nCounter® analysis system or quantified by next-generation sequencing (NGS) where the entire plate can be pooled into a single tube, purified, and sequenced. The reads are processed into digital counts and mapped back to each ROI generating a
The power of the NanoString approach to Spatial Biology
The field of spatial biology has seen exceptional growth in life sciences companies focused on this field. However, NanoString has been the pioneer in this area.
Our spatial transcriptomics motto is “any target, any region, any sample.” There are an estimated 5 million FFPE slides in countless repositories, each carrying intact morphological and molecular information. The data extracted through spatial transcriptomics may support everything from cancer research to new breakthroughs related to infectious diseases.
Imagine how much knowledge one can obtain by interrogating any region of these tissue samples for any biological target. Spatial biology supports biomarker discovery and effective research related to pathways and druggable targets.
Researchers have the power to unravel the information hidden within each and every sample. All you need is one slide and the GeoMx Digital Spatial Profiler. Welcome to true spatial transcriptomics!
Spatial Biology FAQs
What is spatial biology?
Spatial biology is the study of molecules in a two-dimensional or three-dimensional context. Using spatial biology techniques, the users can visualize molecules in their unique contexts within individual cells and tissues. Learn more »
Why is spatial biology important?
Spatial biology is important because it allows the user to study how molecules interact in a three-dimensional setting. Since the body also exists in three dimensions, spatial biology techniques give a more precise view of molecular processes in cells and tissues. Learn more »
What is spatial profiling?
Spatial profiling is the combination of two molecular biology techniques, high-plex gene expression analysis and immunofluorescence. Spatial profiling leverages both technologies to spatially resolve gene expression. For instance, one can use immunofluorescence to identify a region of interest, then collect all expression transcripts in that area. Learn more »
Why is spatial profiling important?
Spatial profiling is important because at the molecular level, form implies function. Understanding how gene expression works in a three-dimensional context is important for a fuller understanding of molecular and cellular biology. Learn more »
What is spatial sequencing?
Spatial sequencing is viewing a specific area or a region within a tissue, then sequencing all transcripts found in that region. Spatial sequencing allows the user to map variation in gene expression across a tissue or region of interest. Learn more »
What are the advantages of single-cell sequencing?
The advantages of single-cell sequencing are that it allows the users to look at specific, unique, or rare cell types within their specific biological context. Previous bulk sequencing experiments that generalized across tissues resulted in the loss of important signals. With single-cell sequencing, individual cells can be chosen and studied based on the research question. Learn more »
What is the difference between single-cell RNA-Seq and RNA-Seq?
The difference between single-cell RNA-Seq and RNA-Seq is workflow. In standard RNA-Seq, all cells from a tissue are maintained in one sample, and these transcripts are sequenced. This gives the user an overview of transcriptional dynamics in a tissue. In single-cell RNA-Seq, cells are sorted first, then specific cells of interest are sequenced. Learn more »
How does spatial sequencing work?
Spatial sequencing works by combining immunofluorescence with next-generation sequencing. Samples are stained with fluorescently labeled antibodies or probes. Using a spatial biology setup, the user selects a region of interest, then uses ultraviolet light to release molecular tags. These tags are then processed using next-generation sequencing. Learn more »

Related Content