Breast Cancer: Best practices for using GeoMx® Digital Spatial Profiler
It’s October and breast cancer awareness month. Breast cancer kills more women than any other type of cancer. It isn’t just limited to women either – a man’s lifetime breast cancer risk is 1 in 833.
A non-trivial amount of federal money is invested into breast cancer research. In fact, compared to other cancers, breast cancer research received far more funding from the National Cancer Institute than other forms of cancer. One area of research involves using spatial biology.
Spatial biology is studying tissues or cells within their microenvironment in the body. This is particularly important when we think about cancers. Breast cancer can be highly heterogeneous. For example, there can be variability between tumors within a single patient, as well as between patients. Understanding how cancer differs at the molecular level can aid in biomarker discovery and identification of drug targets
How can researchers use spatial biology to understand the molecular diversity of breast cancers? One solution is to use NanoString® GeoMx® Digital Spatial Profiler (DSP). To help advance the field of spatial biology to understand breast cancer, specifically using the GeoMx technology to identify biomarkers, a group of researchers called the GeoMx Breast Cancer Consortium wrote a paper detailing best practices for using this technology.
In this blog article, I’ll introduce the GeoMx® DSP technology and give a summary of the recommendations made by the GeoMx Breast Cancer Consortium. Although I’ll do my best to provide a comprehensive summary, please refer to the original article for specific details that go beyond the scope of this blog article.
How does GeoMx Technology Work?
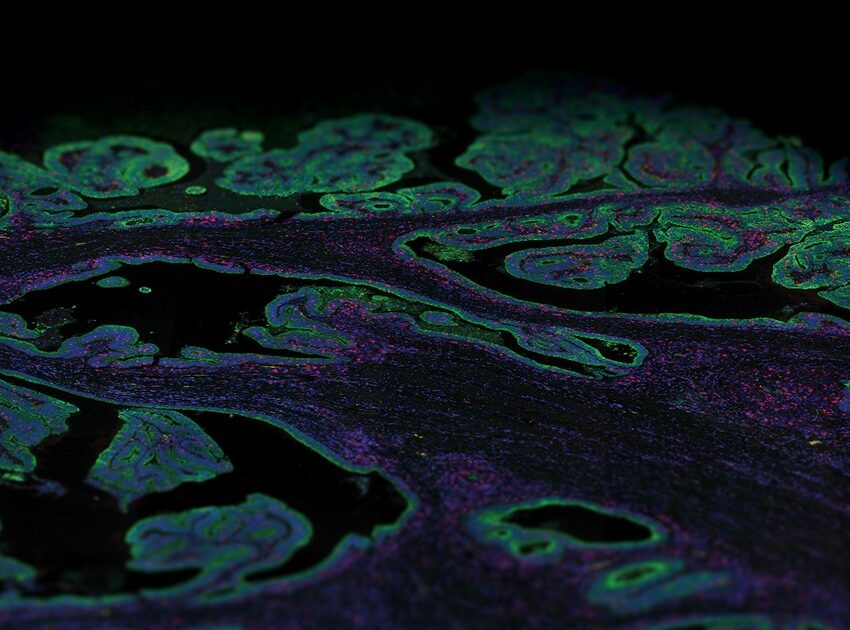
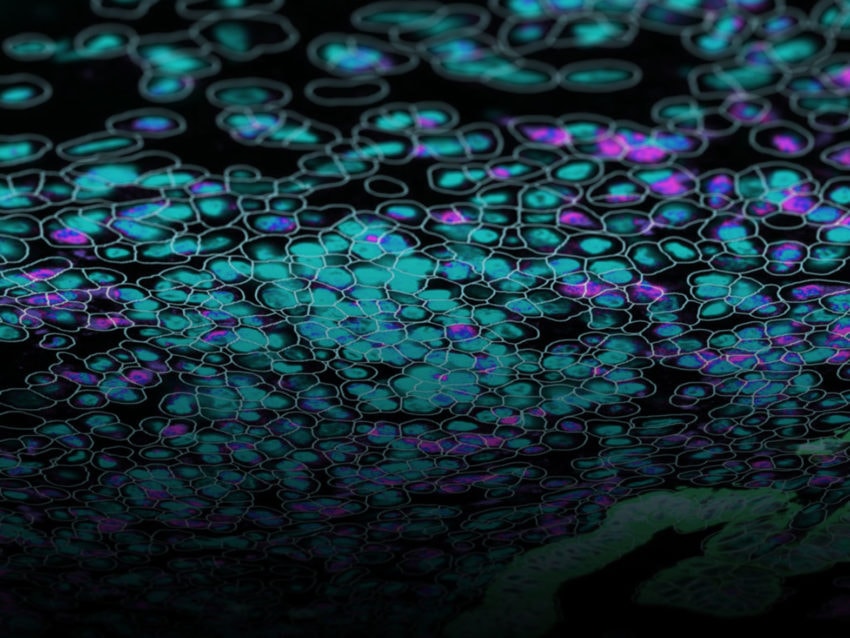
The power of the GeoMx DSP technology is that it allows researchers to capture both spatial or morphological information and gene or protein expression information from a single sample. Additionally, researchers can start with FFPE (Formalin-Fixed Paraffin-Embedded) or frozen samples
First, the tissue sample is stained with either fluorescently labeled antibodies or RNA probes. Antibodies will detect specific proteins within a sample, whereas the RNA probes will detect particular nucleic acid sequences of interest.
Once staining is complete, the GeoMx instrument scans the slides and the user can select regions of interest based on morphological or structural features and their research questions. A UV light is shone onto the region of interest, which causes molecular tags on the antibody/probe to be released. These molecular tags are then collected and counted using either Next Generation Sequencing or NanoString’s nCounter®.
In the context of breast cancer, GeoMx DSP can answer a wide variety of clinically relevant research questions. For example, a researcher could compare gene expression across the tumor and the tumor microenvironment.
Researchers could compare gene expression profiles before, during, and after cancer treatment. This could lead to the identification of novel biomarkers that scientists could use to understand the response to treatment. GeoMx DSP could also be leveraged in healthy adults to understand gene expression profiles in normal tissue.
Best practices for designing experiments using GeoMx
A good experimental design yields good data. Using a sensitive technology like GeoMx DSP can provide unparalleled insights into breast cancer biology – but only if set up carefully.
Given the importance of spatial location, the authors stress that it is important to include a skilled breast pathologist on the research team. A breast pathologist can help with region of interest selection both for the experiment at hand and also facilitate collaboration with other investigators. Alternatively, researchers may also choose to utilize archival images stained with conventional stains, like hematoxylin and eosin.
The authors also suggest that when selecting a region of interest for study to ensure there is an adequate number of cells present. This will make certain that enough sample is collected to process through sequencing. The authors recommend collecting at least 50 cells for protein analysis and 100 for RNA, with 3-5 replicates.
Best practices for data analysis for GeoMx DSP experiments
Breast cancer research can be accelerated with large consortia of scientists such as the GeoMx Breast Cancer Consortium. In order to facilitate these consortia, it is essential that standard data formats are used with capturing and analyzing data. The authors recommend using OME-TIFF and MIAME/MNSEQE compliant data to ensure maximal data sharing.
Other practices recommended by the authors to ensure usability across researchers include the inclusion of standard reference samples across studies. These can also be useful for the normalization process. The authors also encourage researchers to use publicly available toolkits like QuPath to help align methods.
In addition to using standard file formats, the authors encourage researchers to use specific standards for processing the data. For example, for data normalization, the authors specifically recommend the use of housekeeping genes for normalizing small protein or RNA panels. Researchers may also choose to normalize their data using a negative control, such as with IgG staining. For RNA profiling, the authors recommend using trimmed mean of M values (TMM) for normalization.
Since researchers may be studying results across either single or multi-use sites, GeoMx Breast Cancer Consortium authors suggest using normalization approaches that account for both intra- and inter-patient variation. Given the potential for variation, the authors recommend using mixed effect models, rather than looking at averages. For researchers interested in examining pre- and post-treatment data, the authors recommend using analyses such as PCA, tSNE or UMAP to mitigate any potential biases from sampling differences.
Summary
In this article, I explored the science behind NanoString’s GeoMx Digital Spatial Profiler technology. I also summarized the recommendations made by the GeoMx Breast Cancer Consortium for best practices when using GeoMx DSP.
In order for GeoMx DSP technology to be used to the best possible impact, it is important for researchers to adhere to best practices. This includes designing experiments to use the technology to the maximal effect while also ensuring that researchers are measuring what they intend to.
Quality experimental design is important and sets the stage for data analysis. The use of standard normalization techniques and the appropriate data science models is critical for generating data that can be used by researchers around the globe. This will also facilitate future meta-analyses of data collected at many different sites if researchers adopt standardized analysis techniques.
Although this work focused mainly on breast cancer, it is worth noting that the work by the GeoMx Breast Cancer Consortium on breast cancer can serve as a model for researchers interested in other forms of cancer as well. Afterall, approximately 40% of adults in the United States will be diagnosed with cancer at some point in their life.
I’m excited to see how future work with NanoString’s GeoMx Digital Spatial Profiler will translate into exciting new biomarker discoveries and spatial signatures for the development of new targets and a greater understanding of response to treatment.
For Research Use Only. Not for use in diagnostic procedures.